
I am a postdoctoral research assistant in the Hector-Turnbull Lab, starting from 1st Jan 2021. The project is focusing on studying the bias in detecting density dependence in plant communities.
The main aim of this project is to find out how we can measure the relative strengths of conspecific vs. heterospecific competition in an unbiased way. To do this, we will carry out simulation models of plant communities under different conditions and then analyze the resulting data.
The first goal is to decide on suitable simulation models that will allow us to explore a plant community with known parameter values. We can then find out how well we can recover these parameter values when ‘data’ is collected from model simulations. There are a number of existing models that we might use, or we might recode new models from scratch. An illustration of a simulation in an explicitly spatial community of two species with density dependence is given below:
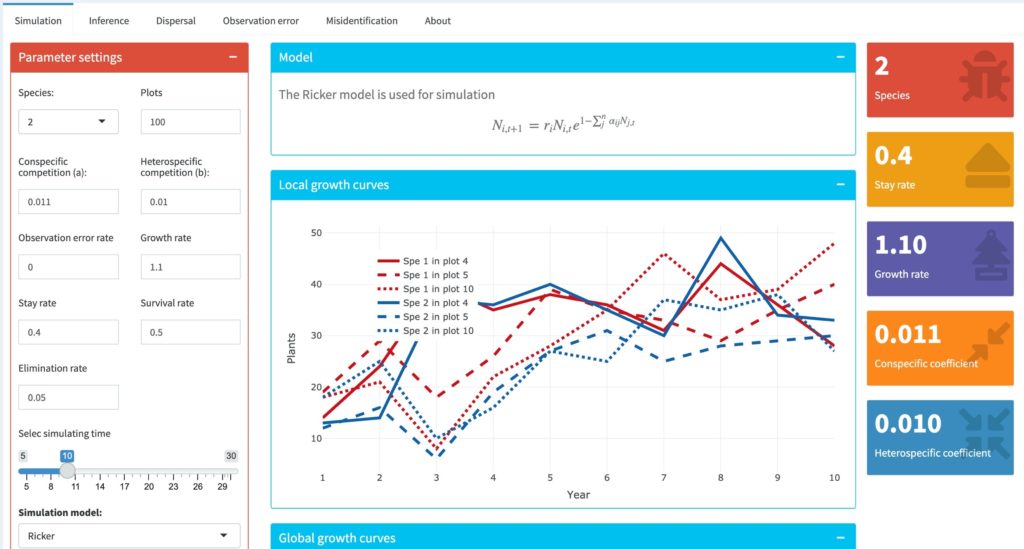
We will explore the bias caused by dispersal, species interactions, observation error and misidentification of competitors, etc. We ask (1) How does the global dispersal affect the local density dependence, therefore have impact on the prediction of the strength of the density dependence? (2) Does the species with higher mobility show strong density dependence or the opposite? Do the heterogeneous morphological traits that relate to the dispersal ability have implication on the strength of density dependence? (3) The underlying interaction mechanisms, such as competition, mutualism, are the direct proxy for density dependence, causing positive or negative covariance among species abundances. However, does different covariance cause different direction of the bias in detecting density dependence? (4) In plant communities, misidentifying rare species is likely to happen due to sampling difficulties in large span of time and space. How does misidentifying competitors cause bias in predicting the strength of density dependence?
A bias in estimating the conspecific coefficient is observed when introducing different magnitude of dispersal:
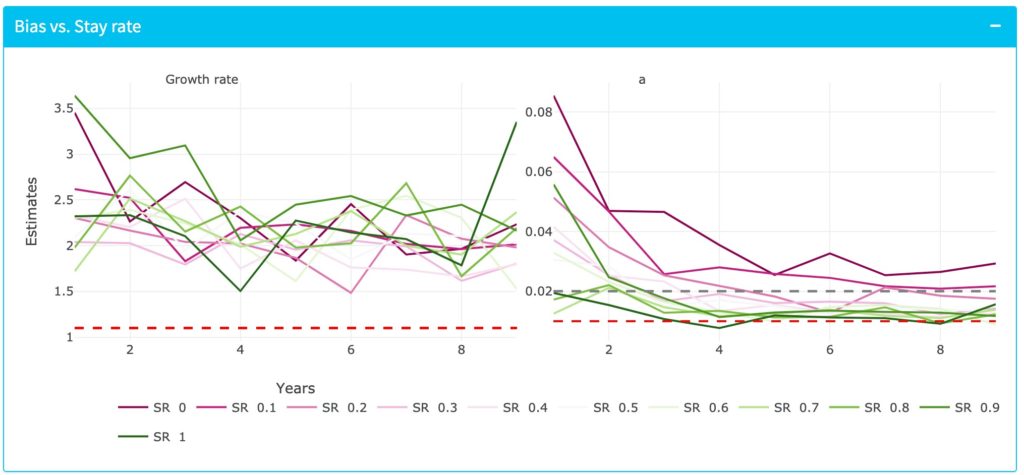
The second goal is to incorporate environmental heterogeneity into the model and see how this affects the outcome. We expect that this will impact our ability to recover the parameter estimates, we will explore potential solutions to this problem that can be implemented in the field.
Previous work
My work is consisting of a variety of theoretical modeling in evolution and ecology.
It fits in two categories:
Eco-evolution
A major challenge in ecology is the need for a better theoretical framework to understand how species assemblages (ecological communities) arise, why some are species-rich and others species-poor, and why some species are present or dominant whereas others are not. My interest is to construct theoretical models, such as stochastic differential equations, to model species interactions in macroecology and macroevolution. Due to the complexity of the biosystem, I also use computer simulation to approach the solution of the puzzle and verify the theory.
Algorithms
Due to the complex system of nonlinearity and high dimension, I invent efficient computational tools like process-based simulations, individual-based simulations, and likelihood-free algorithms to reveal underlying mechanisms from ecological and evolutionary patterns. I am interested in approximate Bayesian computation algorithms that rely on biologically comprehensible process-based simulations. Besides, I am into machine learning algorithms that show advantage on speed and large-scale data processing.
Contact details:
liang.xu@plants.ox.ac.uk
Website
xl0418.github.io
Publications
L. Xu & R. S. Etienne. Detecting local diversity-dependence in diversification. Evolution, 2018 Jun;72(6):1294-1305. doi: 10.1111/evo.13482.
L. Xu, S.Van Doorn, H. Hildenbrandt, R.S. Etienne, Inferring the Effect of Species Interactions on Trait Evolution, Systematic Biology, 2020 Sep; doi: 10.1093/sysbio/syaa072
